PRoBE Laboratory
Pattern Recognition from Biomedical Evidence Laboratory is designed:
-
- To harness prior knowledge for effective knowledge discovery from biomedical data.
- To design and develop novel machine learning algorithms using symbolic, probabilistic and hybrid approaches to solve bioinformatics problems of clinical importance such as biomarker discovery and disease classification.
- To develop complex pattern recognition tools that can be plugged into computer-aided diagnostic systems to facilitate evidence combination from heterogeneous sources such as data from imaging, de-identified clinical information and biochemical profiling.
Research Mission:
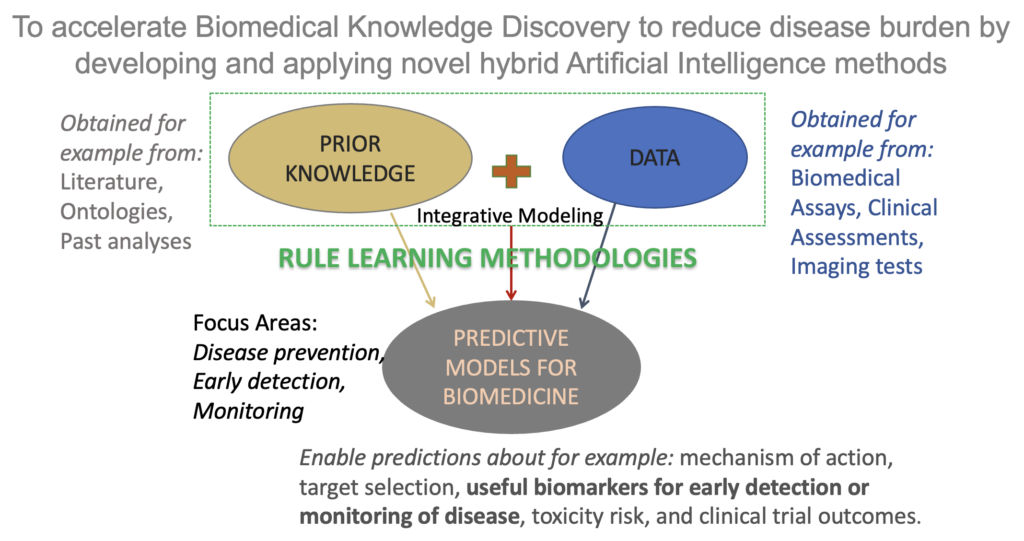
AI approaches to solving real-world problems:
-
-
- Include human in the loop as needed (machine + human combinations)
- Human interpretable predictive modeling (explainable AI)
- Integrative model development from diverse data types and knowledge sources (multimodal collective intelligence)
-
Multimodal Biomedical Dataset Types Analyzed:
- Proteomic
–Mass Spectral (large/wide/deep)
–Immunoassay
- Genomic
–GWAS/SNPs(very large)
–DNA Methylation
–Gene Expression
–microRNA
- Images
–Cardiac MRI
–Brain fMRI
- Microbiome
- Metabolome
- Exposome
- Federated Electronic Health Records (EHR)
Diseases Studied (Biomarker Discovery / Early Detection):
–Amyotrophic Lateral Sclerosis
–Alzheimer’s
–Lung Cancer
–Breast Cancer
–Esophageal Cancer
–Inflammatory Bowel Diseases Ulcerative Colitis, Crohn’s
–Coronary Artery Disease
–Pediatric Cardiomyopathy
–Helminth’s infections
–Beryllium Exposure
–Type 2 Diabetes
–Cardiovascular Disease
–Intervertebral Aging
Grants as a Principal Investigator:
- NIH/ NIGMS K25 Mentored Quantitative Research Development Award: Intelligent Aids for Proteomic Data Mining (5 years)
- NIH/NLM R01 Bayesian Rule Learning Methods for Disease Prediction and Biomarker Discovery (3 years)
- NIH/NIGMS R01 Transfer Rule Learning for Knowledge Based Biomarker Discovery and Predictive Biomedicine (3 years)
- NIH/NIGMS R01 (Renewal) Transfer Rule Learning with Functional Mapping for Integrative Modeling of Panomics Data (3 years)
- PHDA Grant: CADidME: Coronary Artery Disease intelligent detection via Metabolomic Expression (1 year)
Other Notable Research Activities:
- Investigator: NSF CMU Language Technologies Institute – Biological Language Modeling” (4-5 years)
- Investigator: TATRC/DoD – Bioinformatics core (3 years)
- Co-director of Bioinformatics and Biostatistics Core: NCI Hillman Cancer Center – Lung SPORE (Specialized Programs of Research Excellence) (9 years)
- Inventor: US Patent App. 15/107,444, 2016 – Methods for the detection of esophageal adenocarcinoma
Current Collaborators:

Shyam Visweswaran
Professor

Malar Samayamuthu
Senior Research Scientist

Edith Luhanga
Assistant Research Professor
CMU-Africa
Current Graduate Students:

Roy Joy
(Rafael Ceschin: Primary Advisor)

Aidan Lakshman
(Erik Wright: Primary Advisor)

William Reynolds
(Rafael Ceschin: Primary Advisor)
Past Collaborators:

Robert Bowser
Chief Scientific Officer
Barrow Neurological Institute

Michael Becich
Chairman and Distinguished University Professor

Gregory Cooper
Distinguished University Professor and UPMC Endowed Chair

Ali Zaidi
Drexel University Professor of Surgery and Medicine

Jaime Carbonell
Professor

Jill Siegfried
Professor

William Bigbee
Professor

Ashok Panigrahy
Radiologist-in-Chief

Gwendolyn Sowa
Professor

Nam Vo
Professor

Steve Reis
Founding Director of Clinical and Translational Science Institute

David Boone
Assistant Professor

Makedonka Mitreva
McDonnell Genome Institute
Washington University, St. Loius

Harry Hochheiser
Associate Professor

Blair Jobe
Surgeon
AHN
Featured Post Doctoral Advisees:
Doctoral Student Graduates from PRoBE Lab (dissertations)





